This article explains how you can export sequences from the All Sequences Table, the Cluster Tables and Alignments to a downloadable file. You can export either the underlying sequences of your row selection, the table as a csv/excel file, or both together.
If you are instead wanting to take a subset of sequences from an Annotation Result and recalculate clusters, you can view the main article on Extracting and Reclustering.
When exporting sequences from the Cluster Tables, there are also options to extract representative sequences from the cluster. If you are unsure what a cluster is, please see this article: Understanding "clusters". In addition, you may find the Filter Bar useful for pulling out the sequences that meet certain user-specified metrics.
Jump to:
- Exporting Sequences from the All Sequences Table
- Exporting Sequences from Clusters
- Exporting Sequences from Alignments
- Exporting Sequence Tables
-
Saving an Image of selected sequences
Exporting Sequences from the All Sequences Table
To export sequences from any Biologics Annotator Result document, navigate to the All Sequences Table and select the sequences you would like to export.
In the below example, I have chosen to filter on those sequences that have a Fab Tm above 70°, while also having a closest matching Heavy V gene from the IGH1 family. You can learn more about appending Assay Data to your results here, and using Filtering Syntax here.
Go to Export/Extract > Export Sequences in the dropdown.
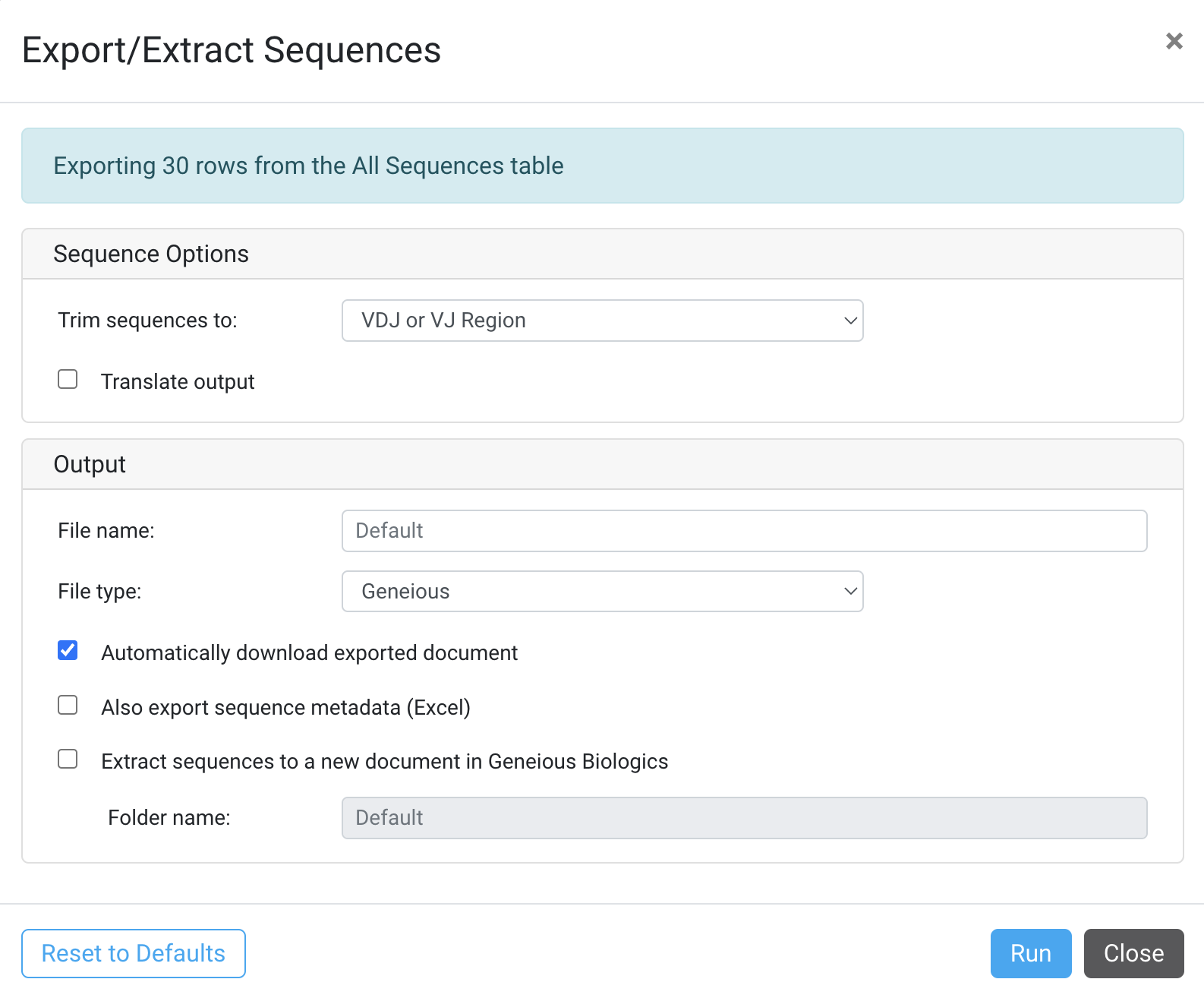
Sequence Options
-
Trim sequences to:
- Entire Sequence (Not-trimmed)
- VDJ Region or VJ Region
- VDJC Region or VJC Region
- scFv Region
-
Translate Output
- Select this to export the amino acid sequence of the selected region
*** Note that if you have Paired Heavy and Light chains, both will be exported from the one row on the All Sequences Table.
Output Options
- File name: Specify the output file name
-
File type:
- Geneious
- Genbank
- fasta
- fasta compressed
- fastq
- fastq compressed
- Automatically download exported document
-
Also export sequence metadata (Excel)
- This will also export the corresponding Table rows representing your sequences as an xlsx sheet. Only the selected rows under Table Preferences will be exported:
- This will also export the corresponding Table rows representing your sequences as an xlsx sheet. Only the selected rows under Table Preferences will be exported:
- Extract to a new document in Geneious Biologics
- Folder Name: Creates a new child folder in Geneious Biologics with the exported document
Click Run to Export the selected sequences.
Exporting Sequences from Clusters
To export sequences from any Biologics Annotator Result document, navigate to your Cluster Table of choice and select the cluster(s) that you would like to export. In the below example the custom cluster for Heavy CDR3 (85% Similarity) was chosen.
In the above image 55 sequences have been found and selected that have the Heavy CDR3 sequence ASYYYGSSSFAY or a Heavy CDR3 sequence at least 85% similar. If you don't know what a cluster is, please see this article: Understanding "clusters".
You can also use the Filtering function to further narrow down your results.
Go to Export/Extract > Export Sequences in the dropdown.
Sequence Options
-
Extract from cluster:
- Extract Representative Sequence (by Liability Score)
- This will extract the highest-scored sequence from within the cluster. Please see this article to learn more about liabilities: Antibody Sequence Liabilities
- All Sequences
- This extracts all sequences contained within the cluster
- Extract Representative Sequence (by Liability Score)
-
Trim sequences to:
- Entire Sequence (Not-trimmed)
- VDJ Region or VJ Region
- VDJC Region or VJC Region
- scFv Region
-
Translate Output
- Select this to export the amino acid sequence of the selected region
*** Note that if you have Paired Heavy and Light chains, both will be exported from the one row on the All Sequences Table.
Output Options
- File name: Specify the output file name
-
File type:
- Geneious
- Genbank
- fasta
- fasta compressed
- fastq
- fastq compressed
- Automatically download exported document
-
Also export sequence metadata (Excel)
- This will also export the corresponding Table rows representing your sequence(s) as an xlsx sheet. Only the selected rows under the All Sequences Table Preferences will be exported (not the cluster table):
- This will also export the corresponding Table rows representing your sequence(s) as an xlsx sheet. Only the selected rows under the All Sequences Table Preferences will be exported (not the cluster table):
- Extract to a new document in Geneious Biologics
- Folder Name: Creates a new child folder in Geneious Biologics with the exported document
Click Run to Export the selected sequences.
Exporting Sequences from Alignments
The Sequence Viewer page of an alignment allows you to select individual aligned sequences and export either:
-
The aligned region
- For example, if a Heavy CDR3 alignment was performed, the CDR3 sequence will be exported
-
The original/parent sequence
- This is the sequence of the original/parent chain used to create the alignment, as it was selected from a Biologics Annotation Result or Comparisons document.
First, select the sequences using the checkboxes and click Export > Export/Extract Selected Sequences. This will bring up the following form:
Sequence Options
Depending on the input for the alignment, these options will be available:
-
Extract selection as:
- Current Aligned Regions
- Original Sequences
- Note: this is not available for alignments from all Table types, but further support is coming.
- Representative Sequence (by Liability Score)
- Note: this is not available for alignments from all Table types, but further support is coming.
-
Trim sequences to:
- Entire Sequence (not trimmed)
- VDJ or VJ Region
- VDJ or VDJC Region
- ScFv Region
-
Translate output (if exporting the original/parent sequences)
Output Options
- File name: Specify the output file name
-
File type:
- Geneious
- Genbank
- fasta
- fasta compressed
- fastq
- fastq compressed
- Automatically download exported document
-
Also export sequence metadata (Excel)
- The associated table rows of the parent sequence will be exported
-
Extract sequences to a new document in Geneious Biologics for editing
- This will create a new sequence list in Geneious Biologics with the output
- Folder Name: Creates a new child folder in Geneious Biologics with the exported document
Click Run to Export the selected sequences.
Exporting Sequence Tables
Before exporting the Sequences Table, you may find it useful to both filter your sequence results and select the columns you want using Table Column Preferences. Below, I have filtered for sequences that have a score above -400 and I have selected to display only the following columns:
- Name
- Chain
- Score
- Heavy CDR1
- Heavy CDR2
- Heavy CDR3
- VDJ Region
*** Note that you can save these column table preferences as Profiles. See this article: How to customise the Sequences Table.
Click Export Table once you have selected your sequences. This will open a pop-up allowing you to select the output format (Excel or .csv) and the option to export only the selected columns, or all hidden columns.
Make sure to select Only Visible if you would just like to export your selected columns.
Saving an Image of Selected Sequences
You can export a .png image of your sequences, and save it in Geneious Biologics for future reference. The image will contain all of the sequences, not just what is in the viewer. It is recommended that you don't export more than 100 sequences at once.
- Select your desired sequences in the All Sequences table.
- Format your sequences as you want to see them in the image. This could be adjusting to the desired zoom level, choosing annotations to show, and adding any data columns in the "Sidebar" that you would like to display.
- Click the "Export as Image" button directly above the sequence viewer.
- Name your image file, and choose whether you would like to automatically download your image. Your image will also be saved in the same Geneious Biologics folder as your original sequences.
