Coloring schemes differ depending on the type of sequence, whether it's a nucleotide or amino acid sequence. The Electropherogram and Quality color schemes are available only for nucleotide sequences while RasMol, Polarity, Hydrophobicity, Clustal and Structural AA color schemes are available only for protein sequences.
To change the color scheme, click Colors in the Sequence Viewer and select the color scheme you wish to use from the dropdown.
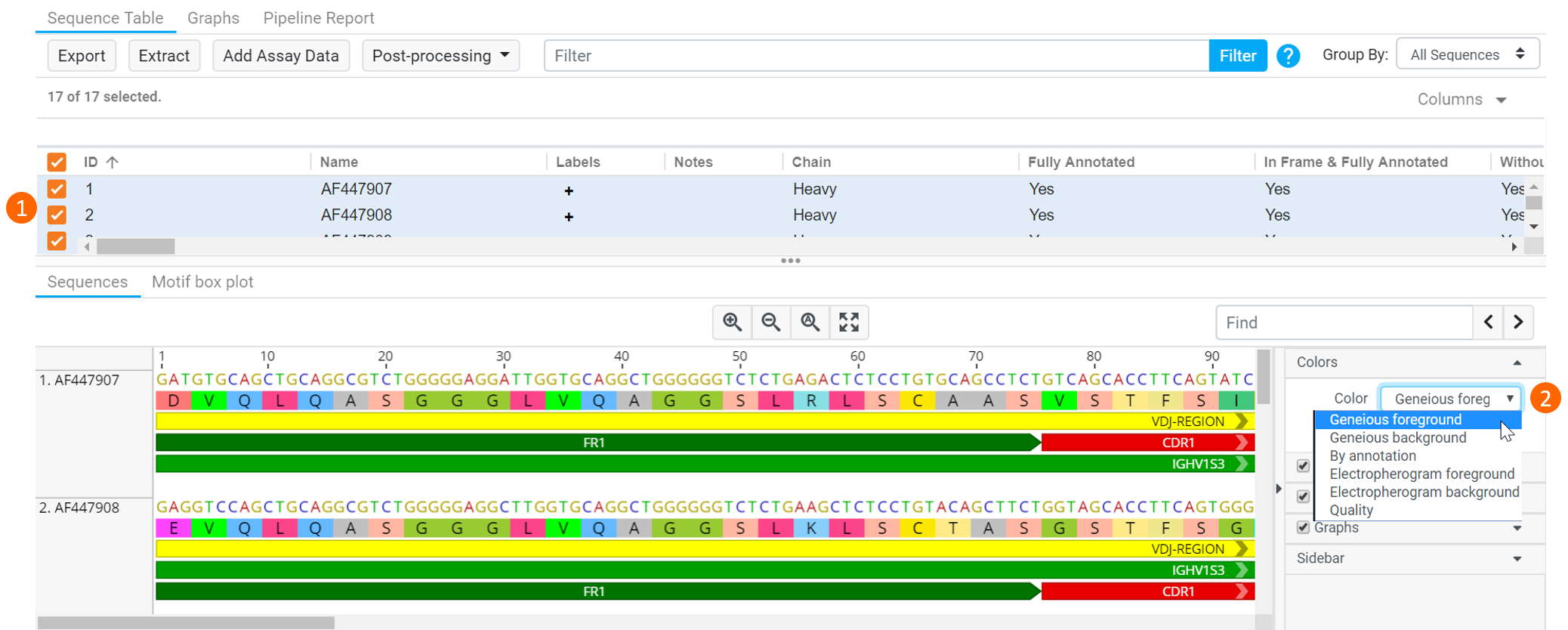
Color Schemes
By Annotation
This colors the nucleotides and amino acids based on the annotation name. In the example below, when Color By annotation and Type FR is selected, nucleotides with the same annotation name and type will share an identical colour.
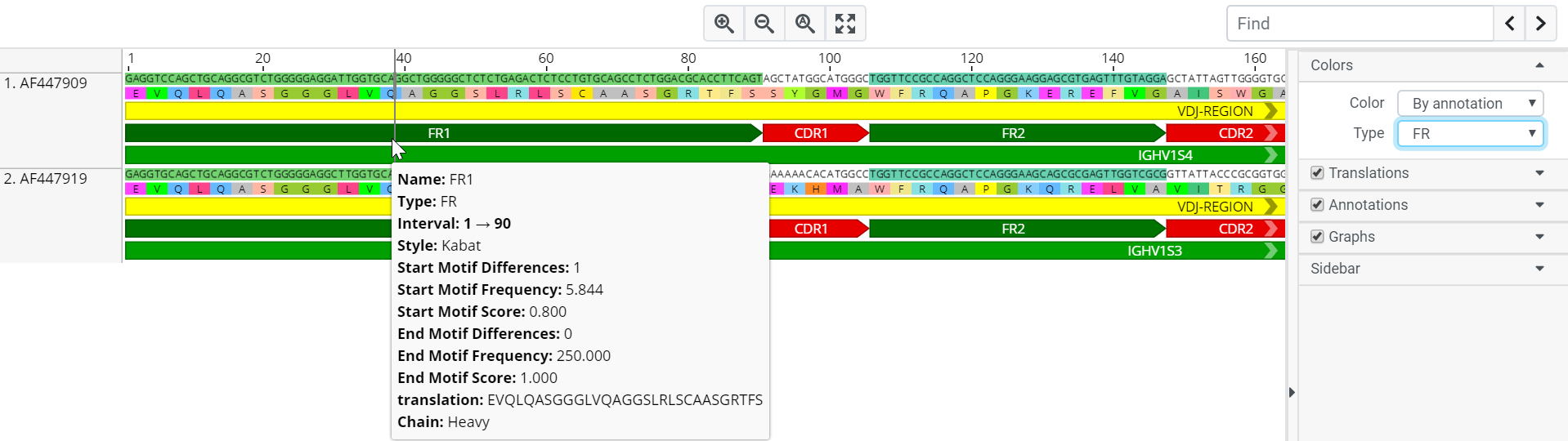
Electropherogram
This colors nucleotides as follows:
Green: A
Blue: C
Black: G
Red:T
Quality
This assigns a shade of blue to each nucleotide base based on its quality score. Dark blue for base quality Phred scores <20,>40. </20,>
Note that when the Quality color scheme is selected for sequences without quality score, the color scheme will default to the Geneious background color scheme.
RasMol
The Rasmol scheme colors amino acids according to traditional amino acid properties. Amino acids associated with the outer surface of a protein are given bright colours and non-polar residues are darker. Most colours are hallowed by tradition.
Bright red: D, Q
Blue: K, R
Mid blue: F, Y
Light grey: G
Dark grey: A
Pale blue: H
Yellow: C, M
Orange: S, T
Cyan: N, Q
Green: L, V, I
Pink: W
Flesh: P
Hydrophobicity
This colors amino acids from red through to blue according to their hydrophobicity value,
where red is the most hydrophobic and blue is the most hydrophilic. The values of the color scale
are given in the figure below. These values are taken from Expasy
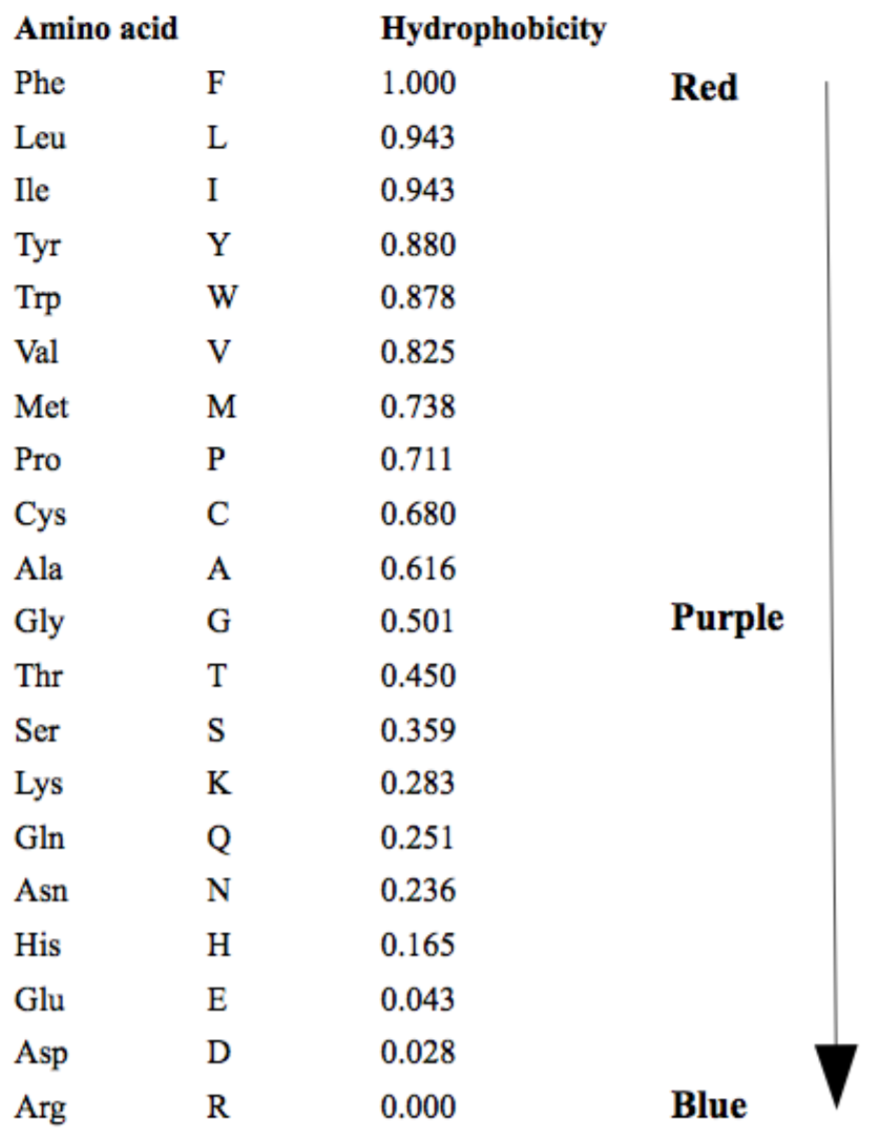
Polarity
This colors amino acids according to their polarity as follows:
Yellow: Non-polar (G, A, V, L, I, F, W, M, P)
Green: Polar, uncharged (S, T, C, Y, N, Q)
Red: Polar, acidic (D, E)
Blue: Polar, basic (K, R, H)
Clustal
This colors amino acids according to their properties and is adapted from Clustal to incude acidic residues as follows:
Orange: G, P, S, T
Red: H, K, R
Blue: F, W, Y
Green: I, L, M, V
Purple: D, E
Structural AAs
This colors amino acids F, Y, W, P and G light green.
