Collections are a way of saving curated lists of annotated sequences, complete with assay data and other relevant sequence information. Collections can be searched against to find any sequences that are closely related to your selected candidates.
There are three ways to add sequences to a Collection:
- Adding sequences when the Collection is created
- Adding and annotating new sequences directly from the Collections page
- Importing chosen sequences from an existing Annotator Result document (this article)
Adding Sequences from an Annotation Result
Sequences can be added to Collections directly from Antibody Annotator, and Single Clone Analysis Scaffold Annotator Results. To do this, open up your Annotated Sequence Result document and then make sure you are viewing the All Sequences table. You can find the Add to Collection option in the Post-processing dropdown. This will only add the sequences selected in the table (up to a maximum of 1000 at a time). It is recommended to only add unique sequences to your Collection.
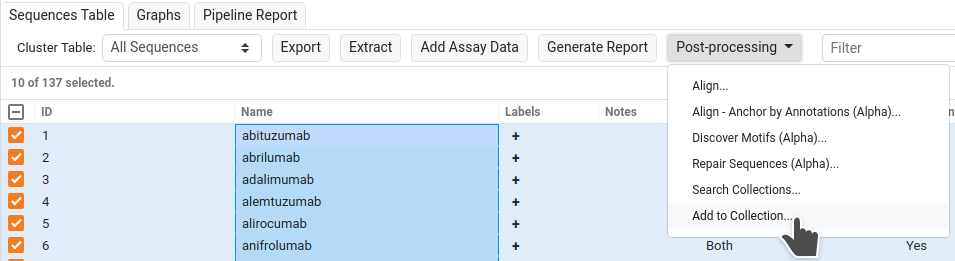
1. Select Collection
There are three stages to the options dialog. First, select the Collection that you'd like to add the selected sequences into.
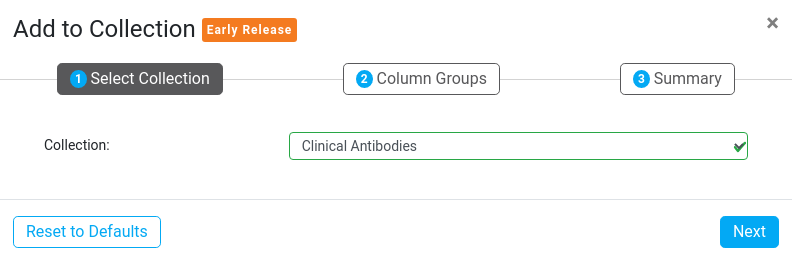
2. Column Groups (Optional)
Column Groups are usually created whenever you add metadata to your sequences, such as imported tabular Assay Data, or File Name schemes. During this step you can manage and merge your assay data groups so that they combine into the expected existing columns in your Collections. This is particularly important if your Assay Data group names have dates in them.
The Column Groups step gives you a list of Column Groups that already exist on your selected sequences that will be imported into the selected Collection. You can choose to either create a new group with a specific name or add to an existing group from the Collection. Adding to an existing group will combine the two group's columns. This will not affect existing row data in the Collection - it will just add on more data below, in the same column.
Add to existing group
If you choose this option, column groups will have their columns merged from both the matching group in the Collection table and the respective All Sequences table the candidate sequences were selected from. Any columns with the same name will be ignored unless they have different types. An attempt to automatically convert the data types will be attempted, but if it isn't possible, then the value will be blank when saved. One example of this is where the duplicate column in the All Sequences table contains words but the column in the Collection table contains numbers. Since words such as "TCG" cannot be converted to a number, then the value will be saved as a blank. The reason columns have data types in Geneious Biologics is to enable filtering and sorting columns in smart ways.
Create new group
Creating a new column group performs just like how the Add Assay Data operation creates a new group, except without requiring a matching column. A new column group with the specified name will be created in your Collection. We don't support duplicate column group names (but you can merge them).
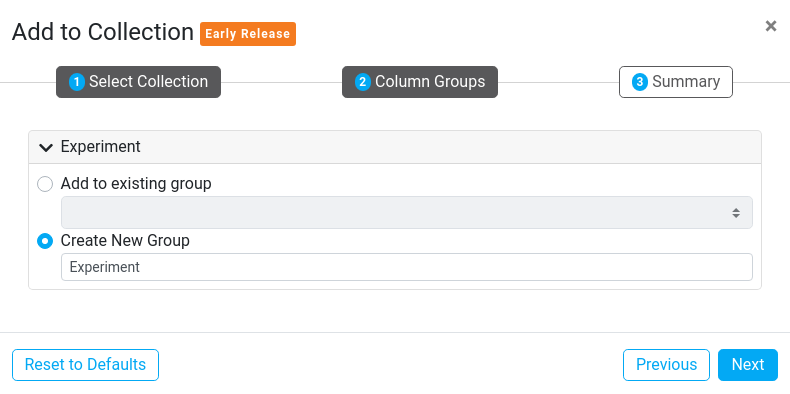
3. Summary
The Summary step shows a brief summary of information from the previous steps. From here you can click the Run button to begin the process of adding the selected sequences to the selected Collection.
What next?
Once the Add Sequences job is complete, your new sequences will appear at the bottom of your chosen collection.
Collections can also be searched from within your analysis Results. For more information about searching for sequence matches, see the Searching a Collection article.
