Over the past few months we have been working on some exciting new features for Geneious Biologics. This includes:
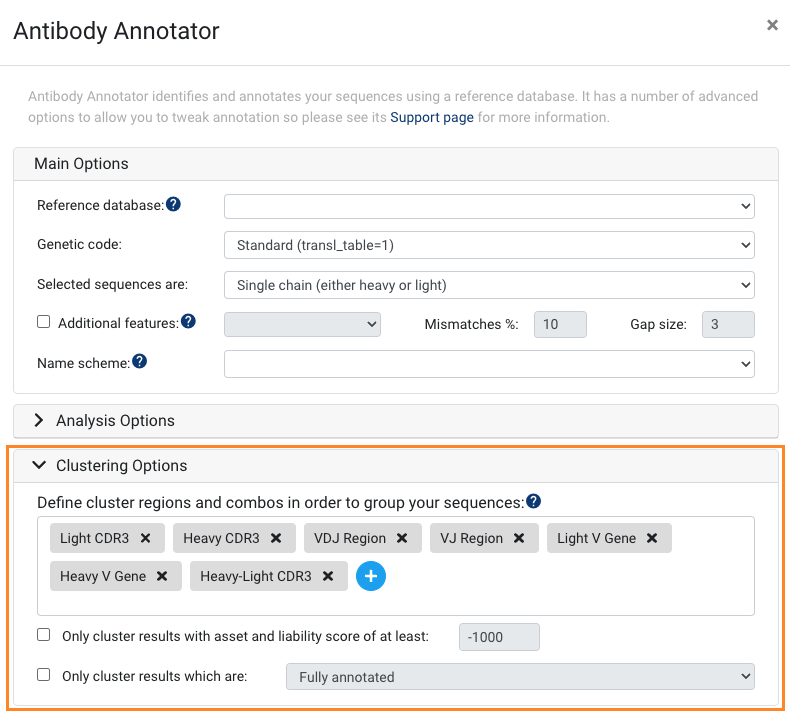
- More user-friendly and customisable clustering options for Antibody Annotator
- Streamlined % clustering in Antibody Annotator
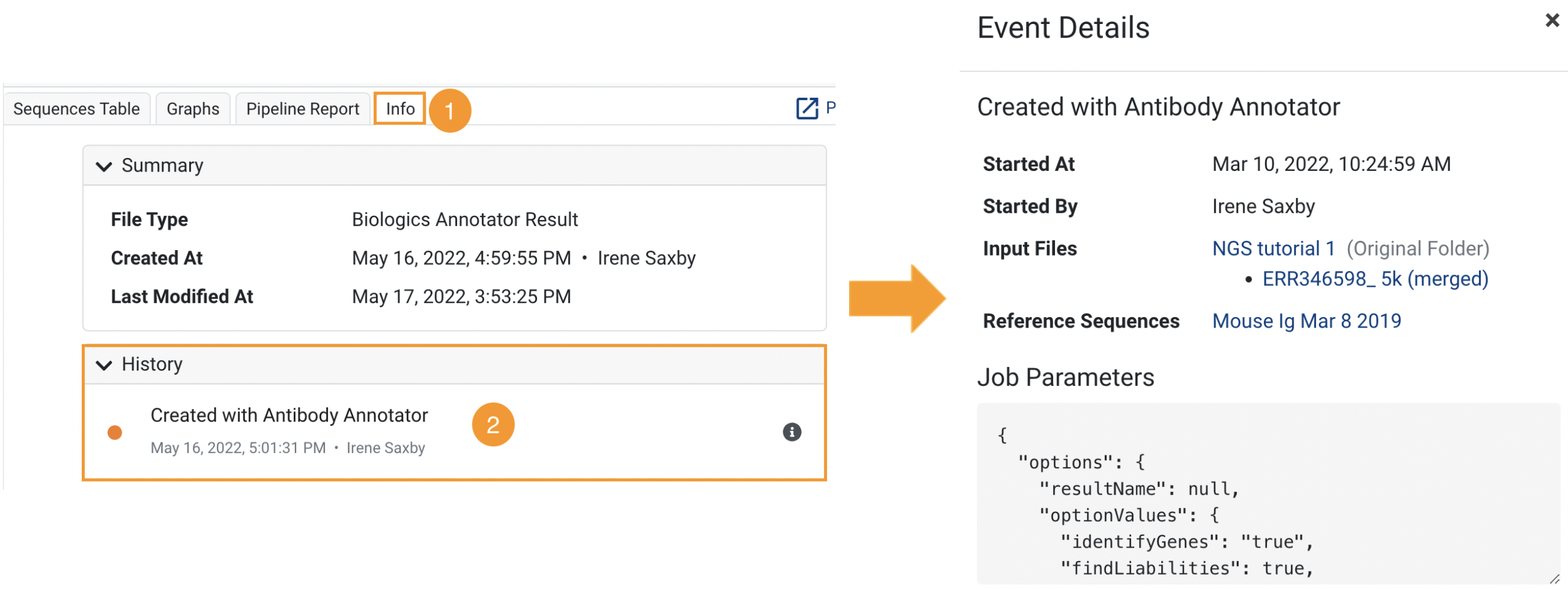
- A new History Viewer tab that allows you to track your input data and more across documents
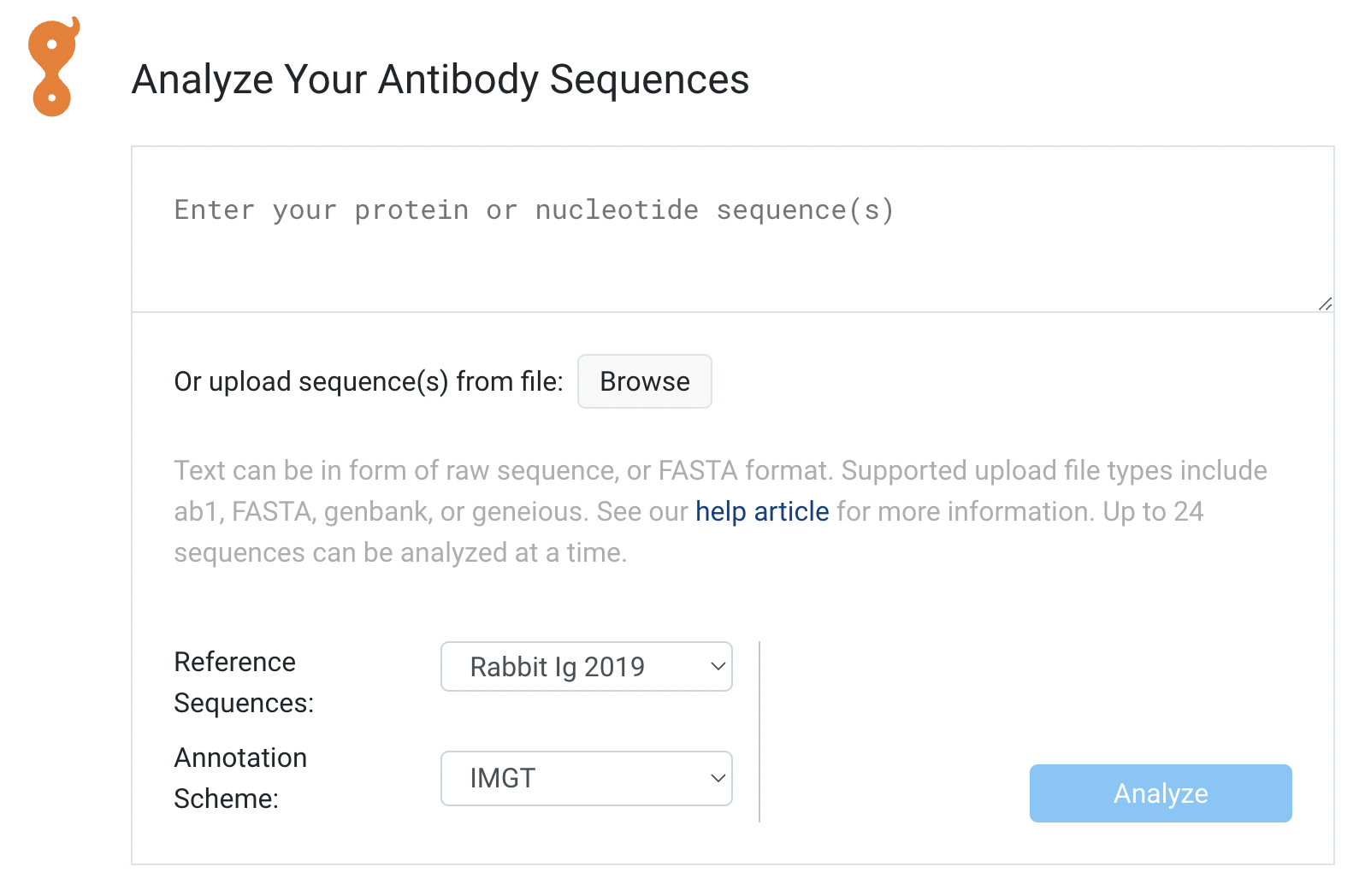
- Copy-paste sequences for Quick Analysis
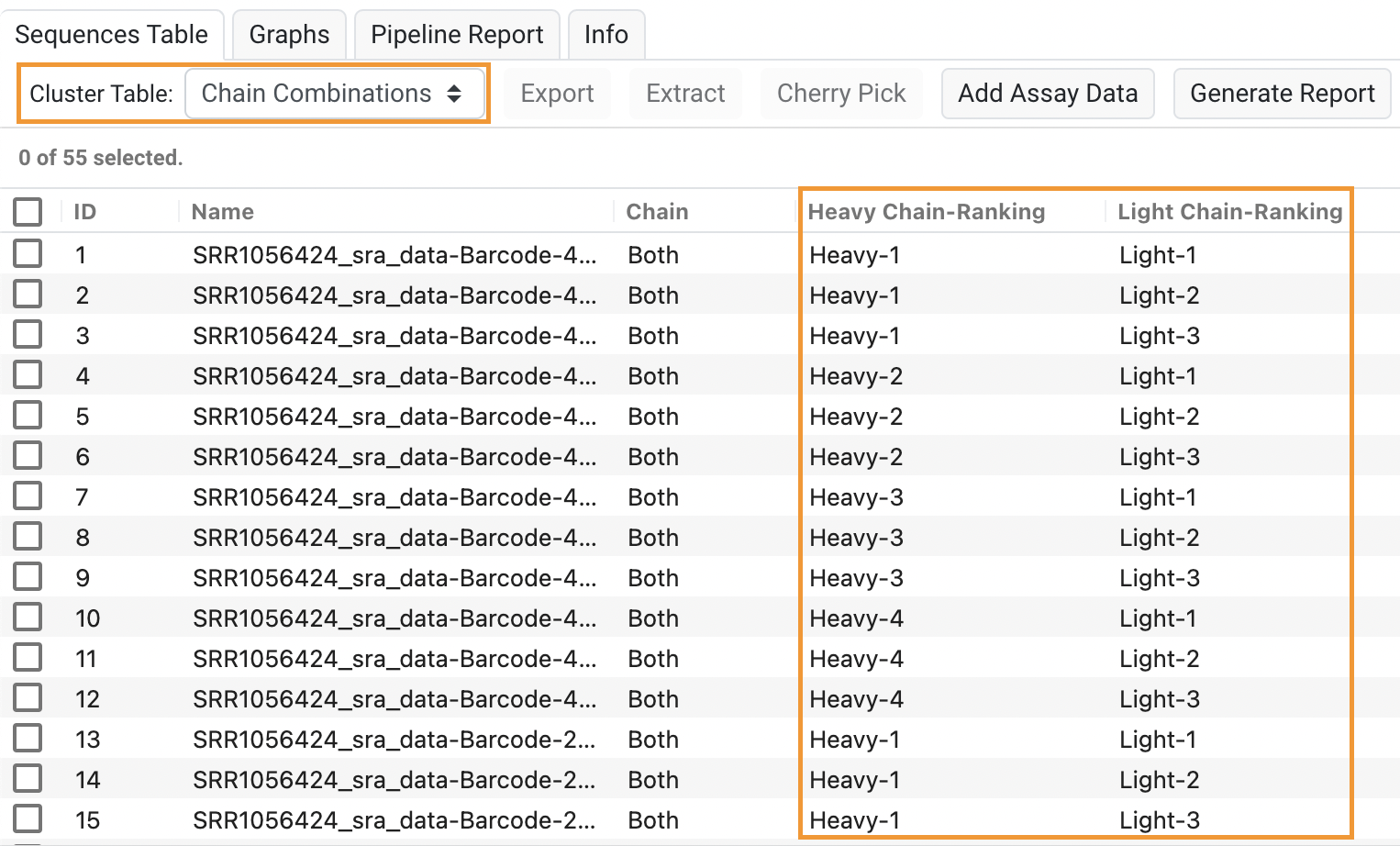
- Chain Combinations tables in Single Clone Antibody Analysis
New Clustering Features
The Clustering Options for Antibody Annotator now look a little different, and allow you to customise clusters right in Antibody Annotator without the need for re-clustering. You can now easily make multi region and gene combination clusters, as well as specify percent thresholds and similarity-based clusters. Please see this article to learn more: Clustering: Advanced Options
History Viewer
Any new analysis result/document produced by Biologics now has extra information tracked in the Info tab under History. This includes which user performed the analysis or uploaded the data, and (if applicable) the job parameters and reference database used.
Quick Analysis
The default home screen when you log in to Biologics is now called Getting Started for premium users. This page has a new Quick Analysis feature that allows you to annotate up to 24 sequences and find the germline genes.
Chain Combinations in Single Clone Antibody Analysis
When running Single Clone Antibody Analysis on barcoded data with a mix of Heavy and Light chains, Biologics will group Heavy and Light chains that have the same barcode together and determine the proportions of different chain pairs within each barcode. This Chain Combinations table is available from the Cluster Table drop-down menu. To learn more about this feature, see the "Understanding the results" section of NGS Tutorial 5. Single Clone Analysis from Separate Lists.