Geneious Biologics (Starter Plan) is a free online analysis tool for annotating and visualizing individual antibody sequences.
If you are using or trialling one of our Premium plans, please see our Premium "Get Started" Guide , as well as our Sanger, NGS, and Single Cell tutorials.
In this tutorial, you will learn how to assemble and annotate raw sequences produced by Sanger sequencing.
This tutorial will cover the following sections:
- Trim ends
- Batch assembly
- Sequence annotation
- Sequence analysis
Get started:
If you haven't seen our Quick Start Guide, we recommend viewing that first.
To start this tutorial, you will need the input data. You can load our example data automatically in Geneious Biologics, by clicking "Load Example data" on the "Getting Started" page. This may take a few moments to run. Alternatively, you can also download the input sequences here and then uploading them into Geneious Biologics.
Sequence trimming
Trimming low quality ends of sequences is normally performed before assembling a contig. This is because the noise introduced by low quality regions and vector contamination can produce incorrect assemblies.
In this exercise, you will learn how to trim low quality bases from both ends of chromatogram sequences. Trim the poor quality bases off the ends of the sequences by selecting all the chromatograms in the Input data folder, then, click Pre-processing > Trim End.
Select the Error Probability Limit option located under Trim By Quality and click Run. This option trims bases up until the point where trimming further bases will only improve the error rate by less than the limit (see image below).
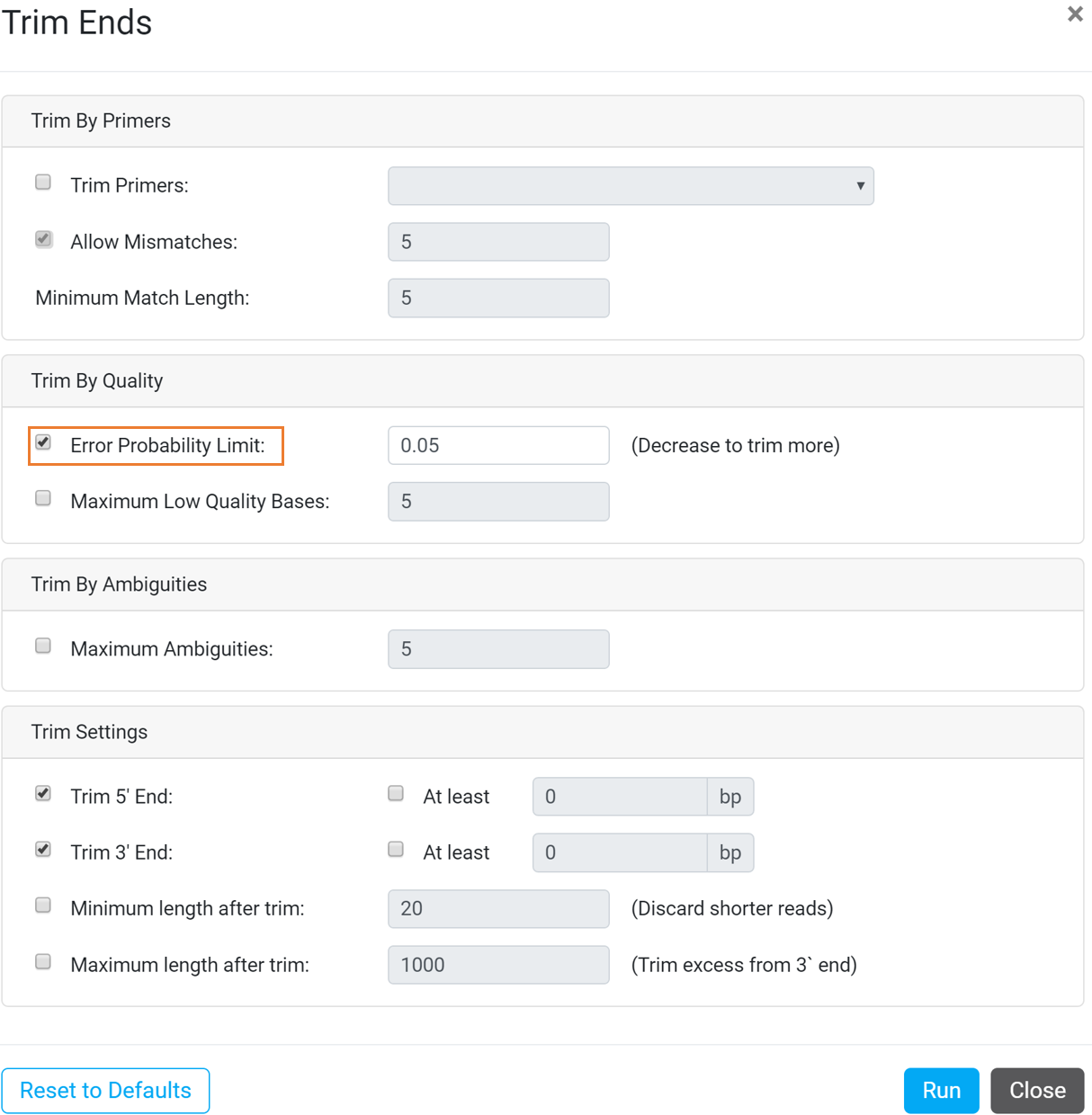
This will produce 6 documents in the Trim ends folder where the sequences are trimmed and shorter than the original input sequences.
**Note that sequence trimming is not necessary for downstream analysis such as sequence assembly and annotation.
Batch assembly
Sequence assembly is used to align and merge overlapping fragments of a DNA sequence to form contig(s) that can be used to reconstruct the original sequence.
In this exercise, you will learn how to assemble chromatograms (i.e. forward and reverse reads of the same sequence) to form contigs. To assemble Sanger sequencing reads , firstly select all of the sequences in the Trim ends folder then click Preprocessing > Batch Assemble Sanger Sequences.
Select the following options from the Batch Assemble Sanger Sequences dialog box and click Run to start the analysis (see image below).
Select the following options:
Batch by Name
- Name part: 4th
- Name separator: _ (underscore)
Assembly Options
- Consensus: call Sanger heterozygotes > 50 %
- Save list of unused reads
- Generate a contig for each assembly
- Output consensus sequences as list
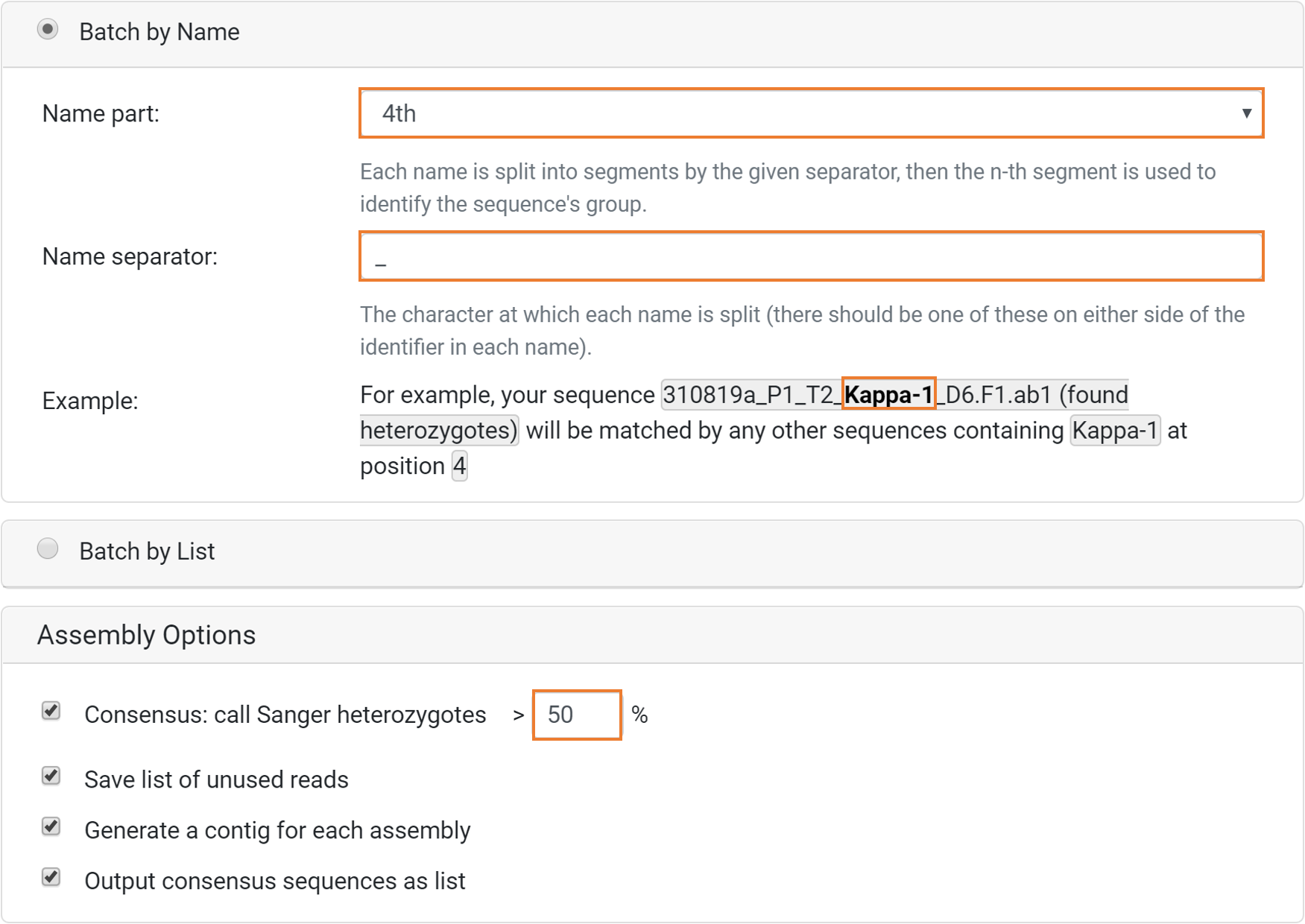
In the example above, sequences that share identical name part when separated by an underscore will be matched together (see Example). Consequently, both the Heavy-1, Kappa-1 and Lambda-1 sequences will be matched together resulting in 3 individual contig files; Heavy-1 Assembly, Kappa-1 Assembly and Lambda-1 Assembly.
**Note that for chromatogram assembly, the orientation of fragments will be determined automatically, and they will be reverse complemented where necessary. Learn more about batch assembly by name and how to assemble chromatograms here.
Sequence annotation
The Antibody Annotator is a versatile pipeline that identifies and annotates IgG-like sequences in reference to an immunoglobulin reference database.
In this exercise, you will learn how to annotate heavy and light sequences. To annotate heavy and light chain assemblies, select the 310819a_P1_T2 Assembly Consensus Sequences document in the Batch assembly folder and click Annotation > Antibody Annotator.
Select the following options from the Antibody Annotator dialog box (see sections and image below).
Input Options
Select the following options:
- Reference database: Human Ig
- Selected sequences are: Single chain (either heavy or light)
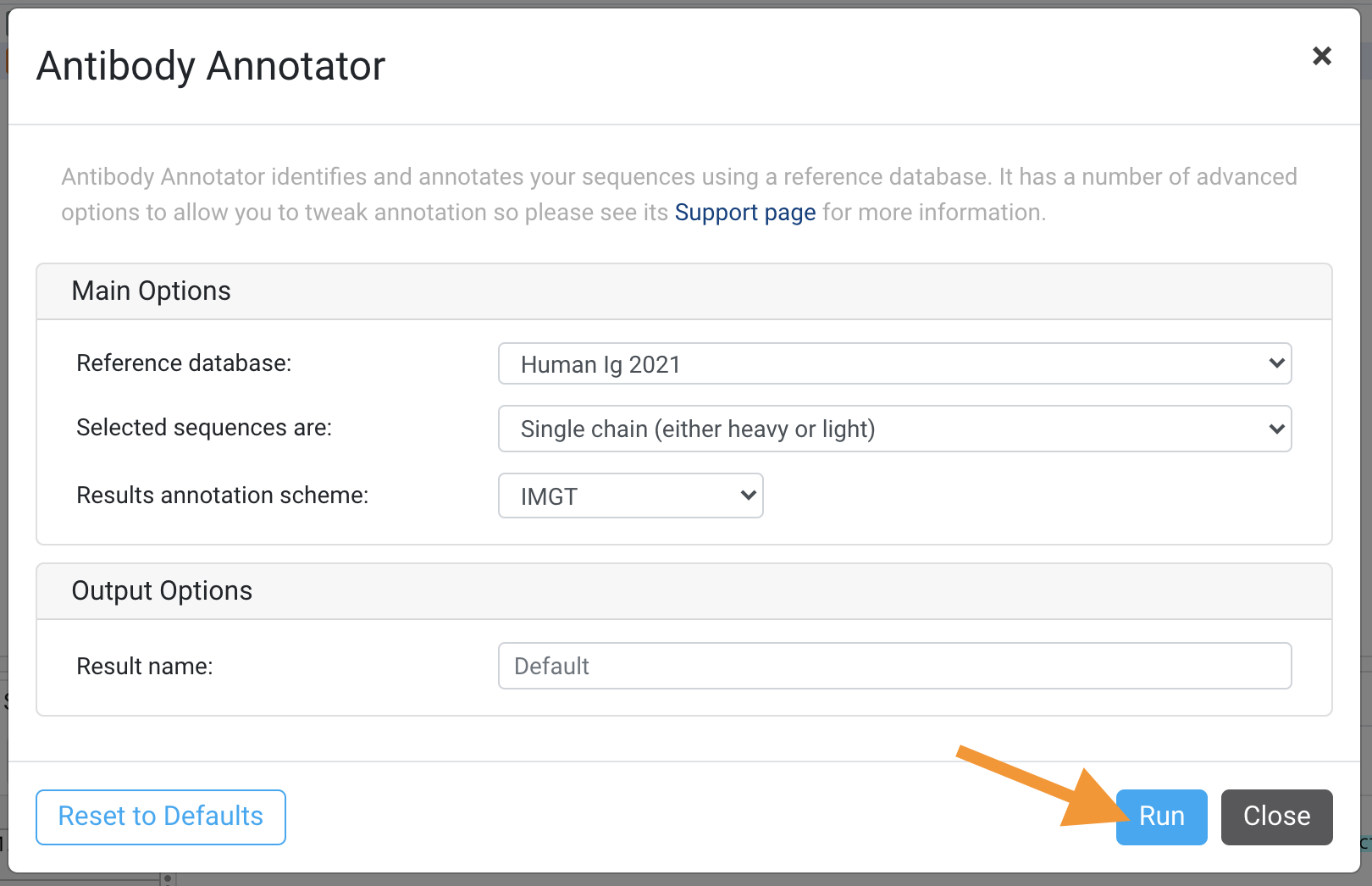
This operation will produce a 310819a_P1_T2 Assembly Consensus Sequences Annotated & Clustered Biologics Annotator Result document.
Sequence and data analysis
This section demonstrates the utilization of the Sequences Table coupled with the Sequence Viewer to analyze sequences. When used together they may aid in rapid candidate selection for downstream analysis such humanization.
The Sequences Table contains details of each individual sequence such as chain type, sequence and region lengths, FR and CDR nucleotide and amino acid sequences, and score to name a few. On the other hand, the Sequence Viewer allows you to view the annotated sequences and search for motifs and annotations.
In this exercise, you will learn how to interpret a Biologics Annotator Result document and search for specific motifs within the sequences. First, select the 310819a_P1_T2 Assembly Consensus Sequences Annotated & Clustered document in the Sequence annotation folder to view the Sequences Table. Subsequently, select Lambda-1 Assembly and Kappa-1 Assembly to view the annotated sequences in the Sequence Viewer.
Lambda-1 Assembly has a distinctly lower liability score compared to Kappa-1 Assembly (-15,230 and 401 respectively) as observed in the Sequences Table and Sequence Viewer (Figure 1.2). The presence of multiple ambiguous bases resulted in a truncated FR2 region.
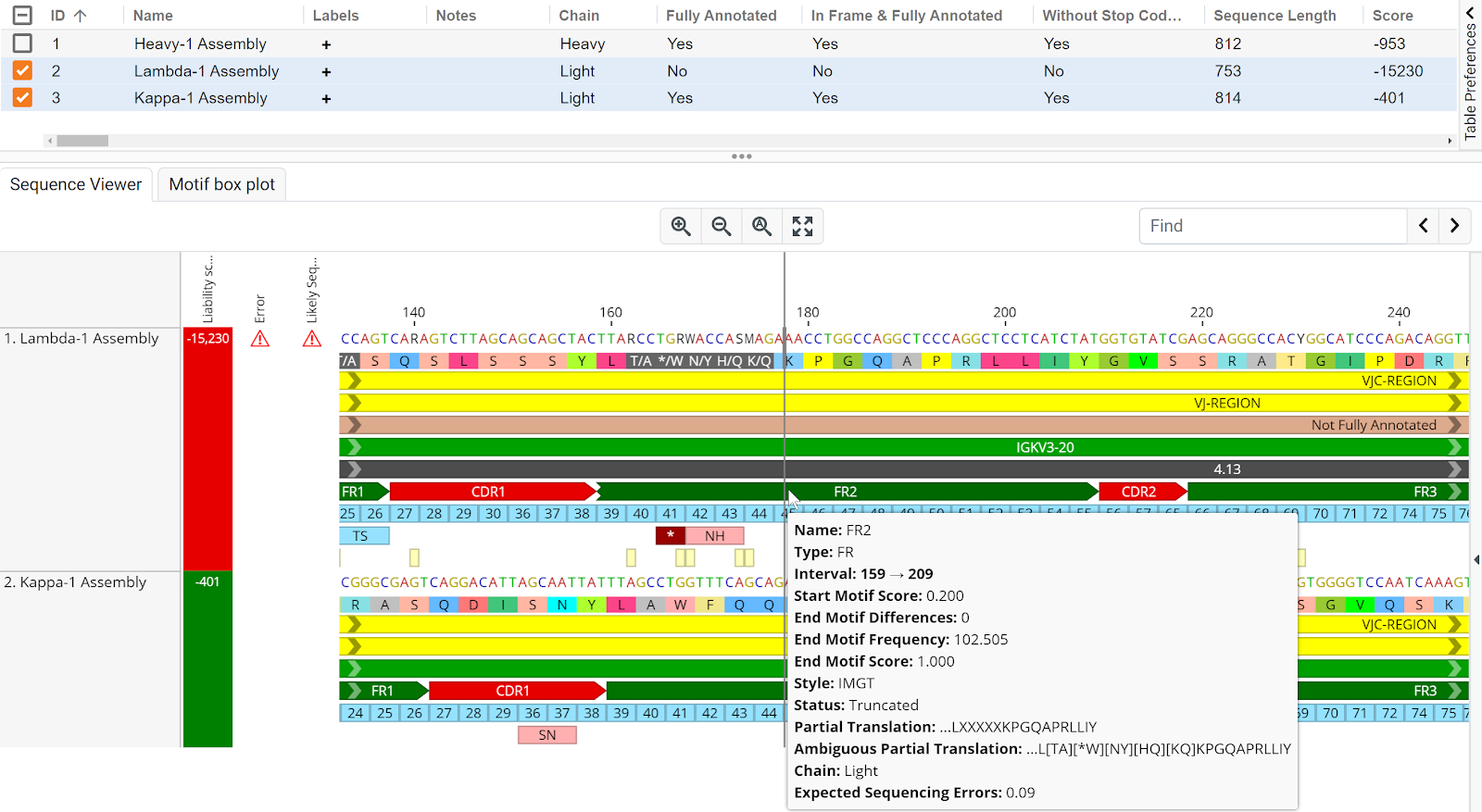
Figure 1.2 | Lambda-1 Assembly and Kappa-1 Assembly sequence annotation. Truncation and the presence of multiple ambiguous bases and a possible stop codon in the FR2 region of Lambda-1 Assembly contributed towards its low liability score.
Heterogeneity may result in base being called as ambiguous bases and this ambiguous bases may affect sequence annotation as observed in the previous analysis (Figure 1.2). To search for ambiguous bases within the selected sequence, simply enter “Type:Contamination” (without the quotes) in the Find text box within the Sequence Viewer and hit enter or ">".
An ambiguous R base is found at interval 105 of Lambda-1 Assembly (Figure 1.3). This contamination annotation is a result of the presence of an alternative base at position 105 of the 310819a_P1_T2_Lambda-1_E7.F1.ab1 chromatogram sequence that was used to generate the Lambda-1 Assembly sequence (Figure 1.3B).
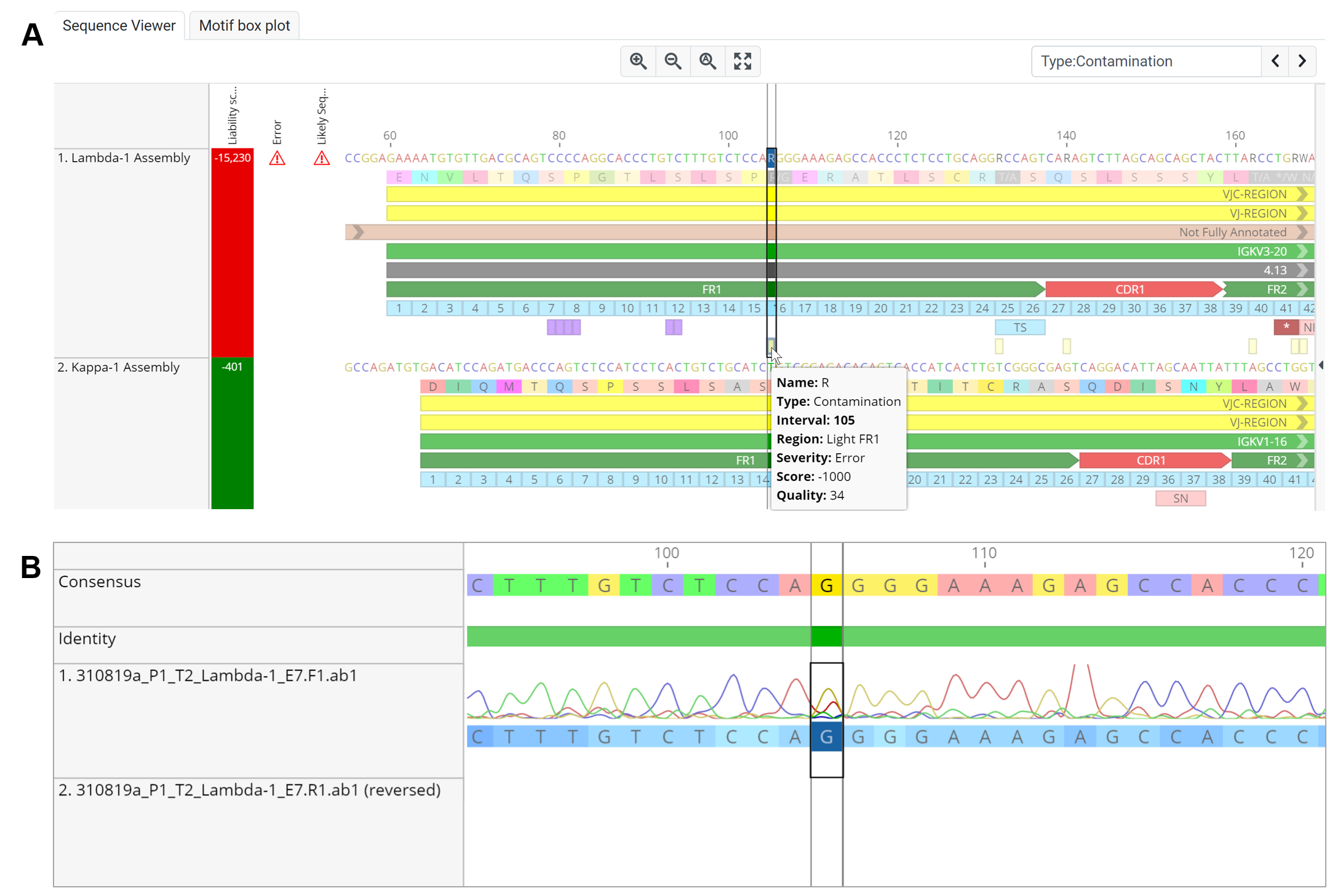
Figure 1.3 | An ambiguous base is detected in the Lambda-1 Assembly sequence. The presence of an A peak that is ≥ 50% the height of the G peak resulted in a heterozygote base being called at interval 105.
**Note that you can also use the Find operation within the Sequence Viewer to search for nucleotide and amino acid motifs. Read more on motif search here.
