The Discovery Motif (Alpha) operation discovers de novo amino acid motifs in sequences or within a region of sequences and outputs a protein sequence list with the motifs annotated onto the sequences. This operation enables quick identification of conserved and common occuring amino acid motifs or amino acid properties making it possible to establish relationships between sequences.
Setting up and running the operation
The Motif Discovery (Alpha) operation can be run within the Files table and Sequence table. Please refer to the sections below to learn more about this operation.
Finding motifs from sequences in the Files table
This operation can be run on nucleotide or protein sequences, sequence lists, or a combination of sequences and sequence lists. When multiple sequence documents are selected, the output for all documents will be combined into a single protein sequence list. If your inputs are nucleotides, then the operation will always translate the input sequences before performing motif discovery.
To find similar or identical motifs in your sequences, (1) select your sequence of interest from the Files table, (2) click Post-processing and (3) select Discover Motifs (Alpha) from the dropdown (see figure below).
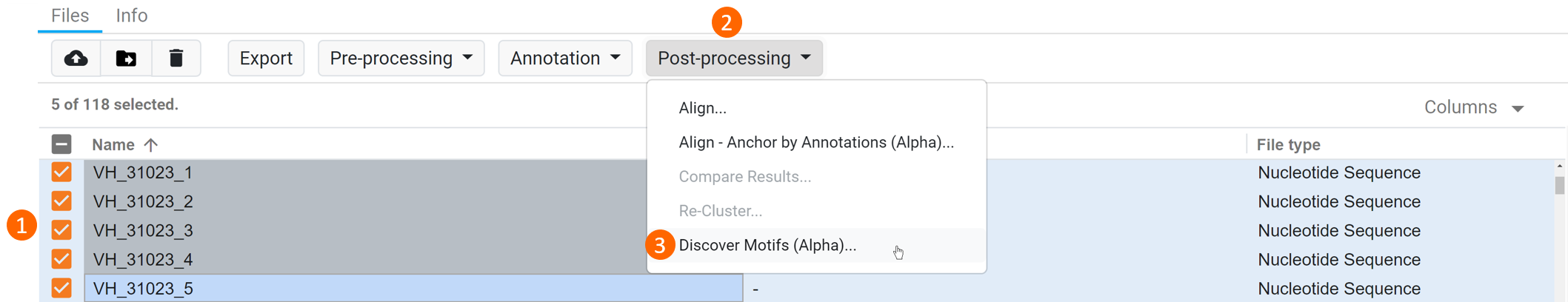
This operation will produce a new Protein Sequence List document and Motif Report. The common occuring motifs are annotated on the protein sequences and not on the original sequences.
Finding motifs from sequences in a Biologics Annotator Result document
To run this operation within a results document, select a Biologics Annotator Results document, (1) select the sequences of interest within the Sequence Table, (2) click Post-processing and (3) select Discover Motif (Alpha) from the dropdown (see figure below).
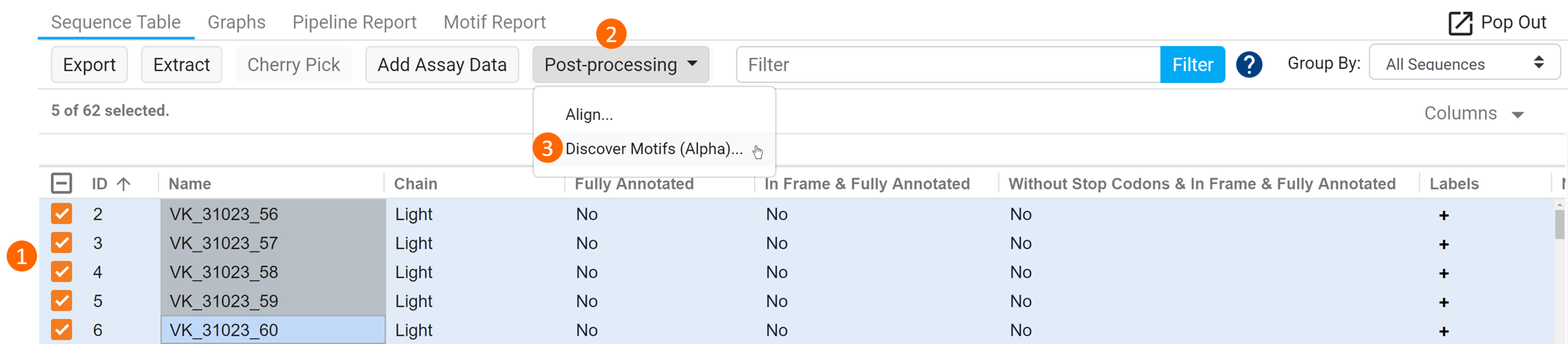
This works the same way as finding motifs from sequences in the Files table but the common motifs are annotated on the original selected result document sequences meaning, the result document is modified rather than creating a new result document. The motifs will be protein sequences, but the corresponding result sequence will still be shown in nucleotides.
Operation outputs
Depending on what the input document is or where the operation was performed, a motif report and motif box plots are produced. Motif annotations remain on the sequence after exporting or other operations, just like a regular annotation would.
Motif report
A motif report is a summary of the motifs found in the selected sequences and is generated for every motif discovery run. To access this report, select the output Protein Sequence List or Biologics Annotator Result document and click the Motif Report tab (see image below).
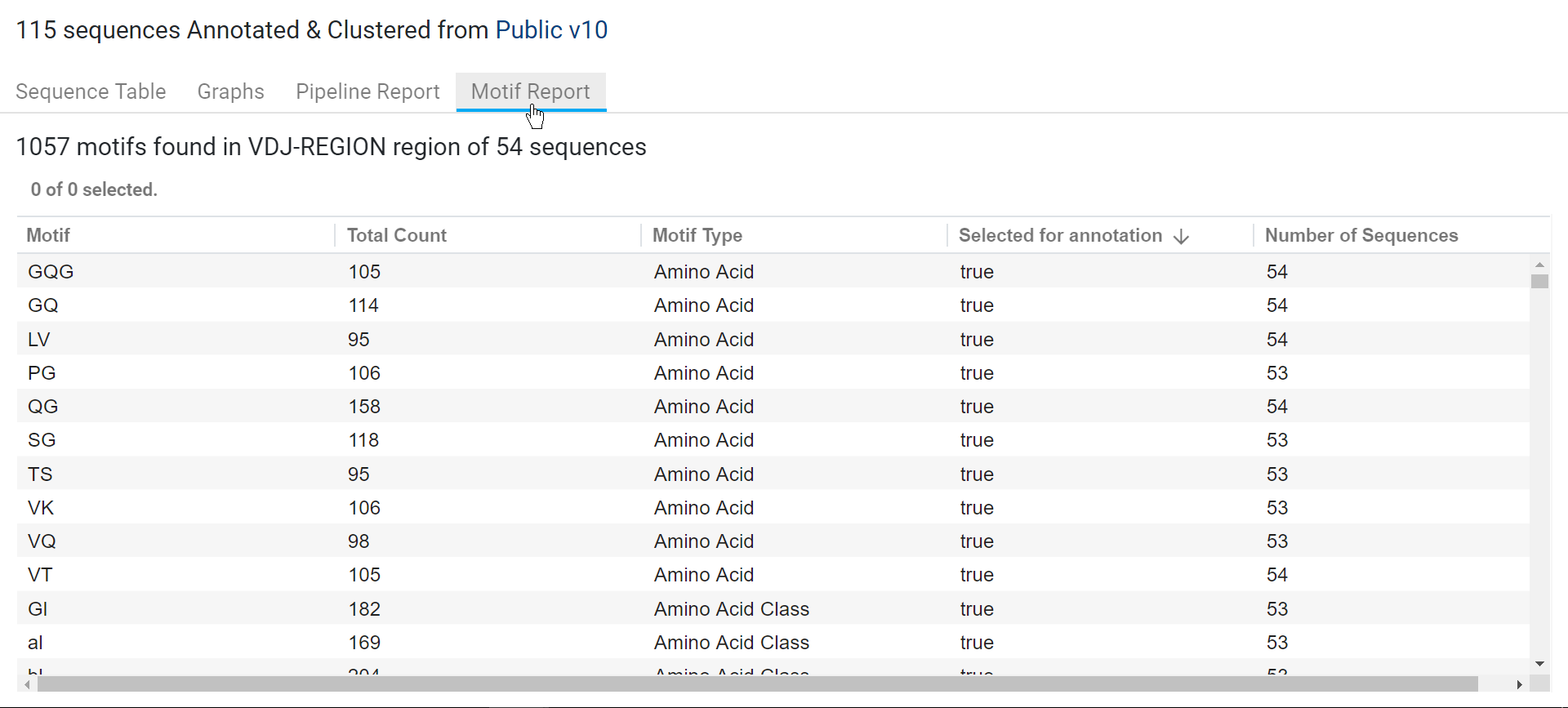
Note that:
- The Total Count is the number of times a motif is found in the all of the sequences selected for the operation. For example, if GQG is found twice in a sequence, this contributes a total of 2 motifs to the Total Count value.
- The Number of sequences is the number of sequences that consist of a motif. For example, if GQG is found twice in a sequence, this contributes a total of 1 motif to the Number of Sequences value.
- Only motifs with "true" values for the Selected for annotation column are annotated on the sequences.
Motif box plot
When the Discover Motif (alpha) operation is run within a Biologics Annotator Result document, motif box plots (whisker plots) are generated on the fly based on the selected assay data values. These motif box plots may be useful in establishing relationships between a motif and assay data. Please refer to the following article on how to add assay data to a Biologics Annotator Result document and the sections below on how to view these box plots.
Viewing motif box plots
Motif Box plots can be viewed in a tab next to the sequence viewer. Two sets of boxplots are generated:
- One set generated from all the sequences that were originally selected for the operation
- One set that represents your currently selected sequence in the Sequence Table
To view the motif box plot in the presence of assay data, (1) click the Motif box plot tab and (2) select the appropriate assay data value from the dropdown to view the motif box plots (see figure below).
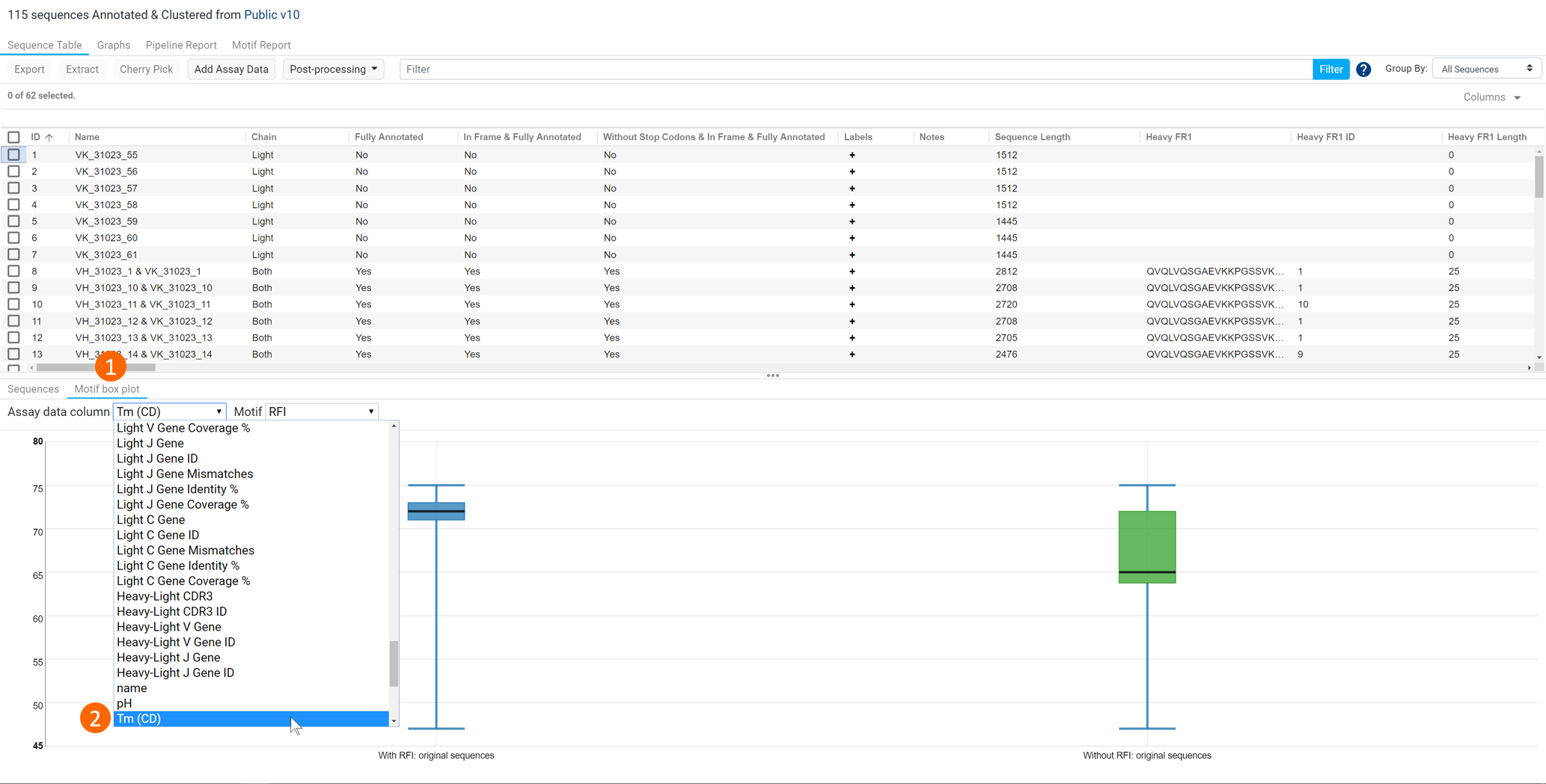
Note that:
- Motif box plots are only generated when the Discover Motif (Alpha) operation is run within Biologics Annotator Result documents.
- Only numerical Assay data columns are supported.
Examples of motif box plots
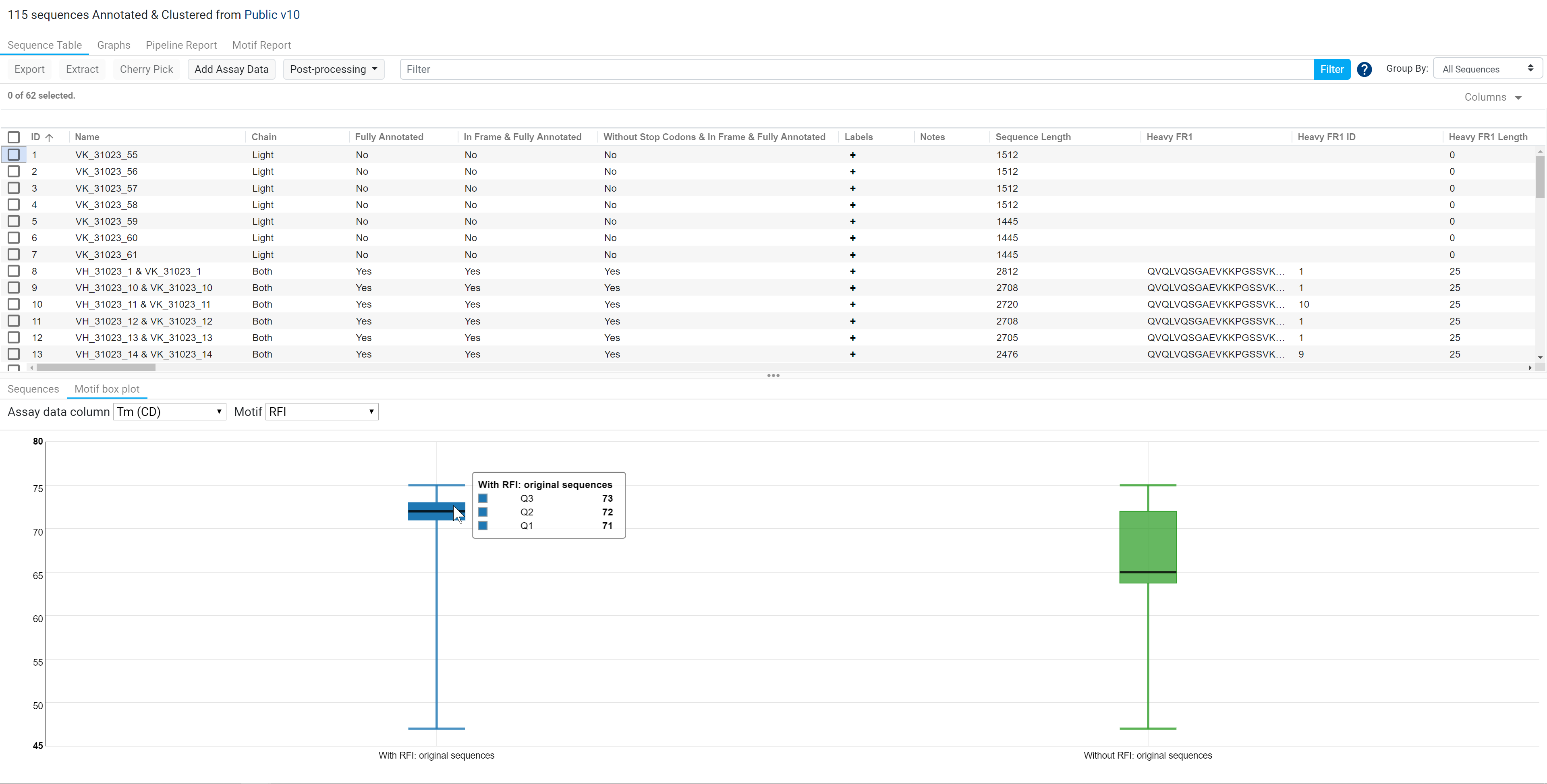
In the example below, when no sequences are selected in the table, the motif box plot will only display the box plots of the correlation between Tm (CD) and Motif: RFI for all of the initial sequences selected for the operation (right box plot: sequences without RFI and left box plot: sequences with RFI). The box plots show that sequences consisting of the RFI motif, have a median melting temperature of 72°C.
In the example below, if your selected sequences do not have any motifs, these sequences will be excluded from the box plots. Likewise if a selected sequence does not have any value for the selected assay data column, it will also be excluded.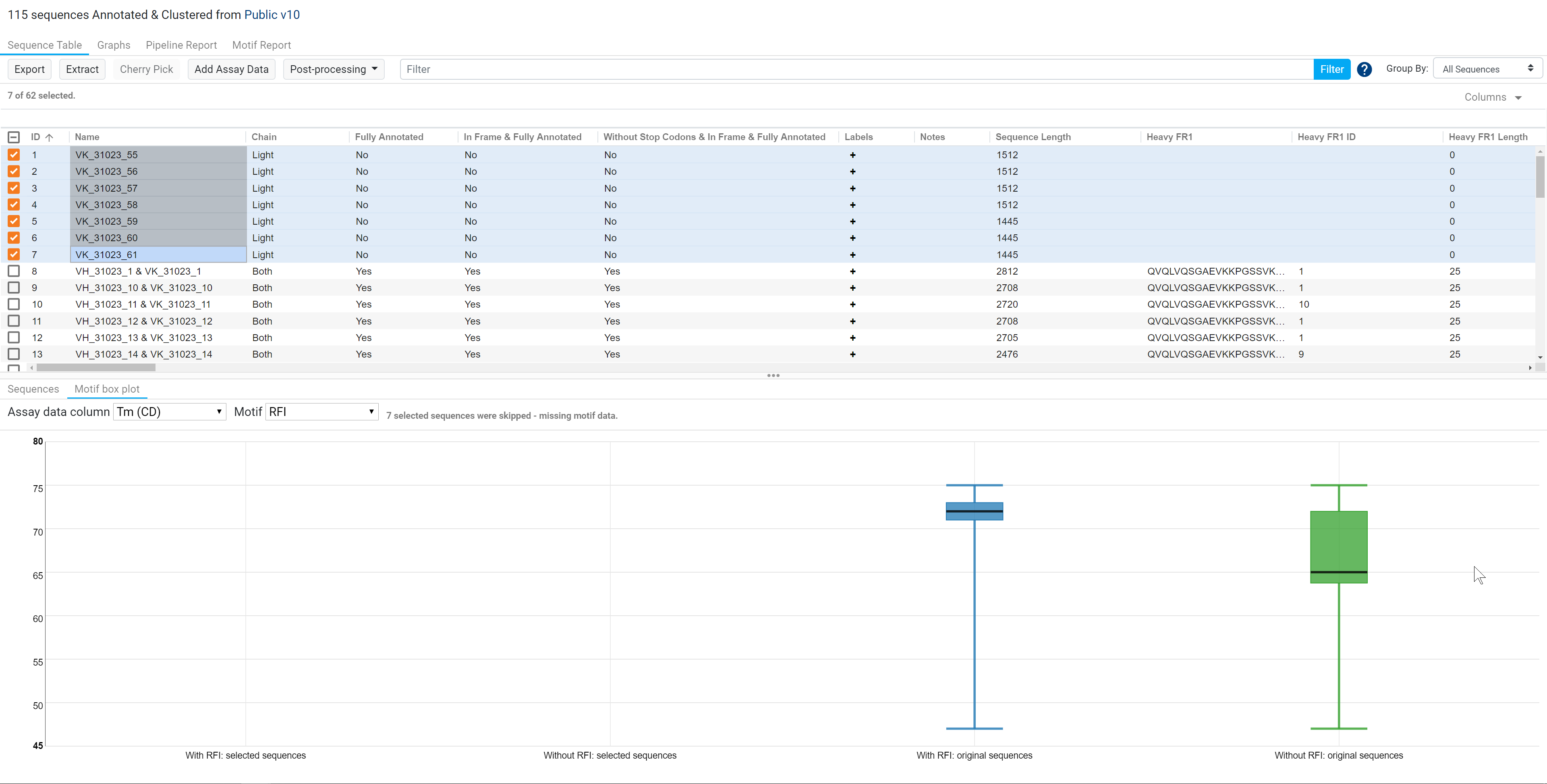
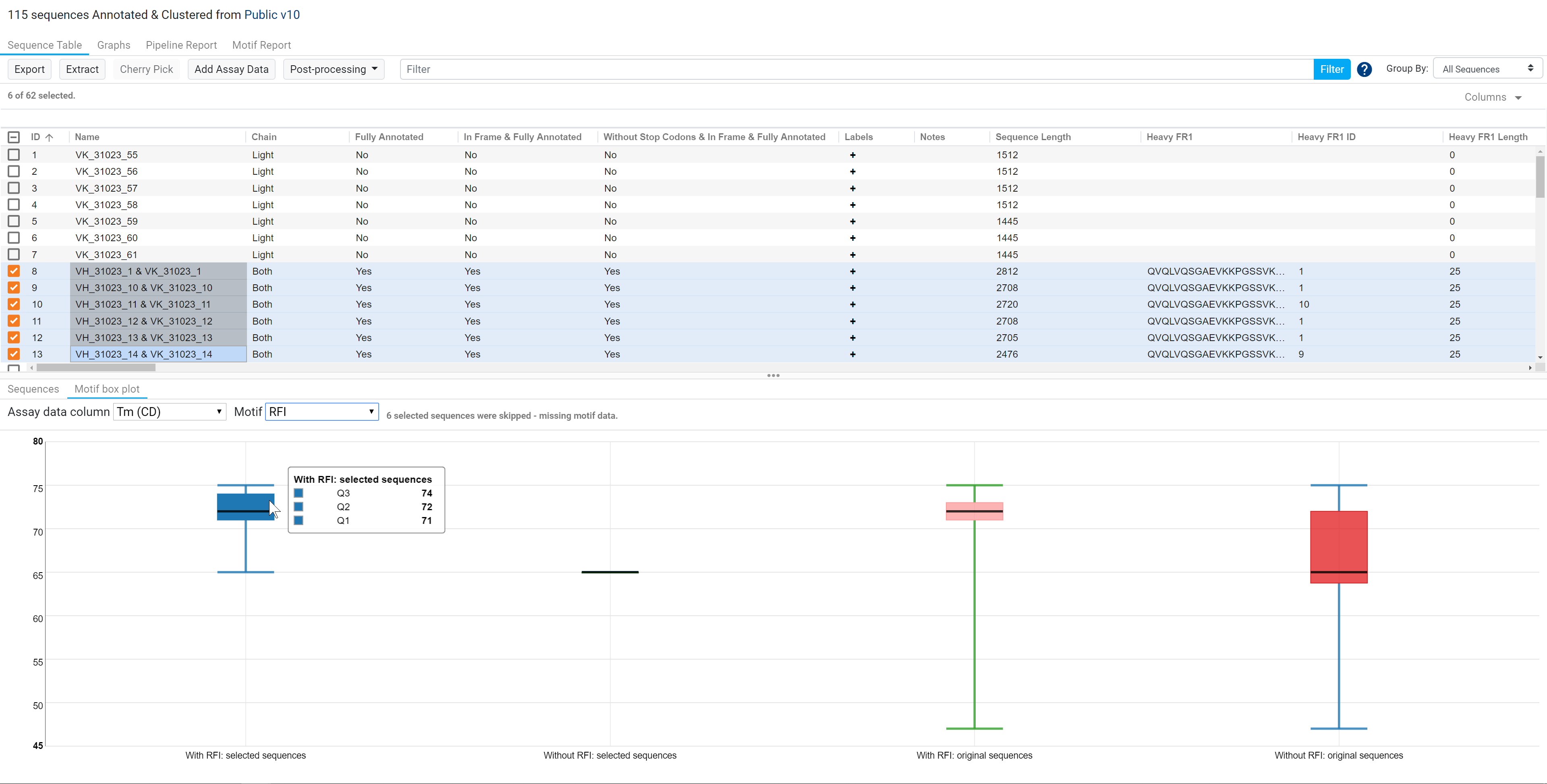
In the example below, out of the 6 paired sequences (6 heavy and 6 light), 6 of the sequences were skipped as they were not part of the originally selected sequences. The median melting temperature for the other sequences that consist of the RFI motif is 72°C while one of the selected sequences that do not consist of the RFI motif has a melting temperature of 65°C.
How the Discover Motif (Alpha) algorithm works
The operation works as follows:
- This operation finds the most common motifs within the selected parameters and a motif that occurs 3 times within a single sequence it is just as "common" as a motif that occurs once in three different sequences.
- If there are more motifs present than the number specified by the user, the motifs are ranked and the top ones selected.
- Motifs are ranked by:
- How "common" they are, then
- Their length with longer motifs ranked higher, then
- Alphabetically based on the motif sequence
- All motifs found are present in the Motif Report, but only "selected" motifs are annotated and available in the Motif Boxplot.
- A subsequence is not considered to be a motif if a longer motif exists with the same frequency. For example, if ASN occurs 7 times, ASNP occurs 7 times, and SNP occurs 10 times, then:
- ASNP is considered a motif as it's the longest.
- SNP is considered a motif, because although it is a subsequence of an existing motif ASNP, it is more frequent than it's parent super-motif.
- ASN is not considered a motif, because it is a subsequence and occurs with the same frequency as ASNP.
The amino classes are defined as follow based on IMGT:
- Aliphatic (l) - Ala (A), Val (V), Ile (I), Leu (L)
- Sulfur (s) - Met (M)
- Hydroxyl (h) - Ser (S)
- Acidic (c) - Asp (D), Glu (E)
- Basic (b) - His (H), Lys (K), Arg (R)
- Amide (m) - Asn (N), Gln (Q)
Single AA classes - Phe (F), Try (W), Tyr (Y), Gly (G), Pro (P)
Important notes
- The operation does not currently work on multi-interval annotations.
- You should not run Motif discovery twice at the same time on the same Biologics Annotator Result document.
- Motif boxplots only support numerical column data at this time.
- If you attach new assay data to a result, you may need to refresh the page before the new columns appear as options in the boxplot.
- If you perform motif annotation a second time on the same result sequences, then the first set of motif annotations will be removed and replaced with your new motifs. If you perform motif annotation a second time on the same result document but with different sequences selected, then the old motifs will remain on your original sequences, and the new motifs will appear on the newly motif-discovered sequences.
- The box plot only shows motifs from the most recent Motif Discovery operation. It also only includes sequences that were selected as inputs for the most recent Motif Discovery pipeline job.
